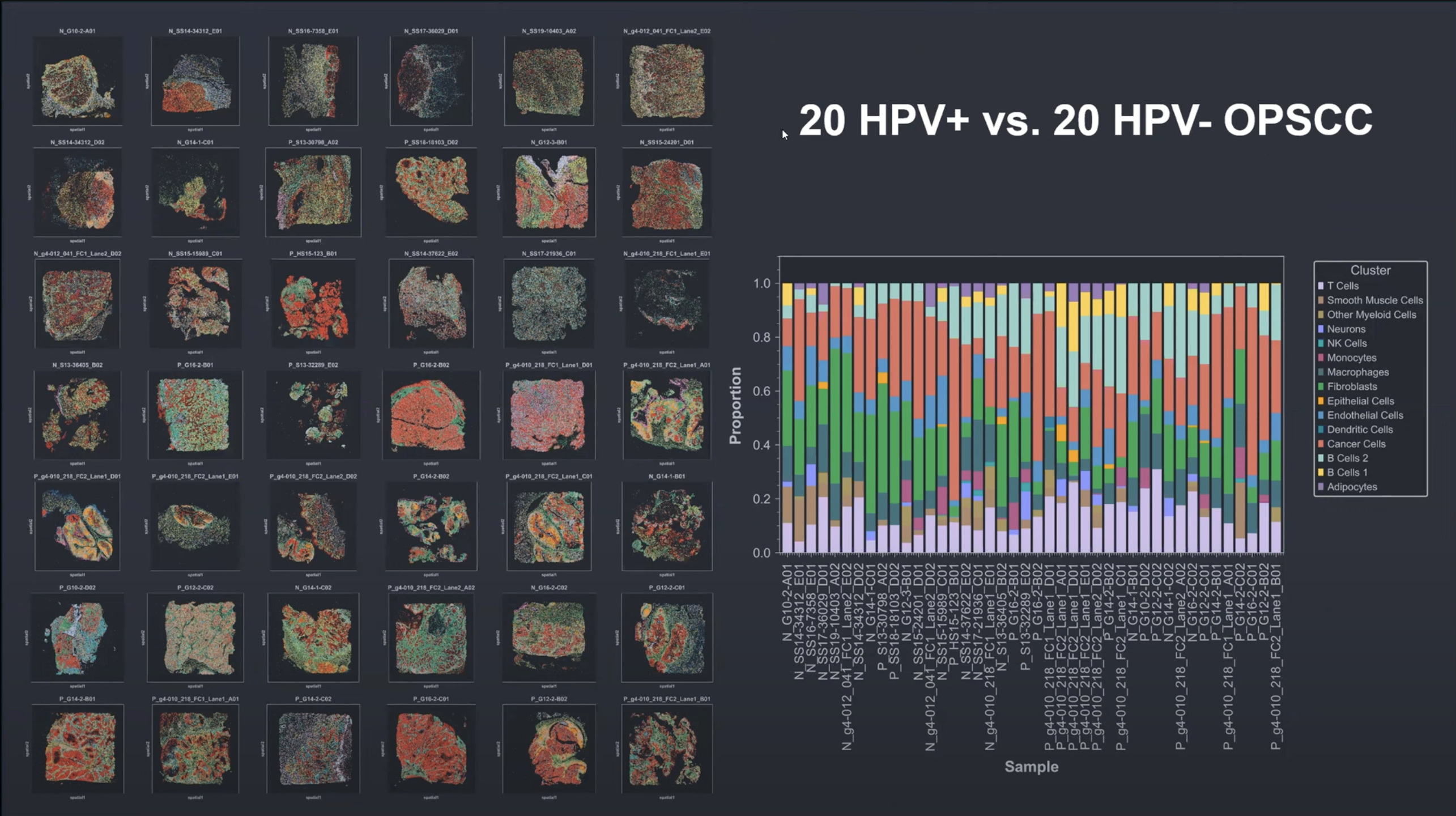
G4X Platform and Ecosystem
In Situ Multiomics with Speed, Scale and Reproducibility for Serious Production.

5-Day Sample-to-Answer
From FFPE section to single-cell spatial multiomics in five days — transcriptomics, proteomics, and fH&E from a single tissue sample.
Tissue Transfer
Patented, Scalable Tissue Handling Methods
- Controlled tissue handling - Minimizing the impact on valuable FFPE tissue while maximizing imaging area usage.
- Reduction in tissue waste - Eliminates the need to score blocks or use tumor micro-arrays (TMAs).
- Efficient storage and transfer of tissues- Users can store tissue on hydrogel pads after sectioning or on slides after transfer.
- User-friendly, high-volume operation- Enables simple transfer of a full run of 128 samples in as little as 3 hours.

Chemistry and workflow
Innovative Chemistry, Optimized for FFPE
- Efficient Capture in FFPE Samples - High affinity, short probes enable effective RNA capture from fragmented archived clinical sample types.
- High Sensitivity for RNA and Protein Detection- Amplification-based detection of padlock probes on target RNA regions or protein.
- Proprietary Methods for Direct Sequencing - Purpose-built technology to enable gap-fill sequencing of variable RNA sequences directly in cells, in tissues (coming soon).
- Streamlined Wet Lab Workflow - Simplified 2.5-day workflow enabled by our purposefully-designed workflow suite.

System Operations
Flexible, High-Throughput System Operations
- Established Instrument- The G4X has a field-matured base platform with years of real-world use and customer insights as a sequencer.
- High-Speed Imaging System- Operating at over 2 billion pixels per second, G4X imaging occurs in a single day, enabling multiple runs per week.
- Flexible-High Throughput Run Capacity - 40 cm2 of tissue imaging, divided across 4 flow cells allows users to adapt the run size to their project needs.
- Independent Lanes - Each flow cell has 2 or 4 fluidically independent lanes, allowing users to run up to 16 panels per run.

Analysis
Powerful Analysis Suite
- Integrated Single-Cell Analysis: The G4X performs base calling, transcript identification, clustering and cell segmentation for robust single-cell results and run QC.
- Multiomic Results Viewer: A flexible web-based platform allows for layering fH&E images, protein images, and transcript localization at subcellular resolution.
- Plug-and-Play with 3rd-Party Platforms- Standard spatial file outputs empower users to leverage their preferred open-source or commercial secondary analysis tools for deeper insights

Panels
Modular, Application-Specific Panels
- Well-Curated and Performance-Optimized - Expert-selected gene sets to ensure each panel is precisely curated to characterize tissue-immune interaction and accurate cell typing.
- Modular Design for Versatility - The flexible architecture allows researchers to combine our refined core immune panel with disease-specific modules tailored to their project needs.
- Customizable for Targeted Insights - Easily add up to 50 genes to a panel, create a new 150-gene module, or create fully customized 500-gene panels to meet specific research goals.
Specifications
G4X Spatial Sequencing Platform
Component
Details
Technologies
In Situ Multiomics
Beyond in situ multiomics, G4X is capable of fast, flexible next-generation sequencing from 4-color rapid SBS chemistry.
Sequencing ?
In Situ Modalities
500-Plex RNA
18-Plex Protein
fH&E
Direct-Seq is a novel approach for in situ sequencing of up to 100 bases of variable RNA sequence in target regions directly in tissue samples. (Coming Soon)
Direct-Seq ?
In Situ Panels
Panel Explorer
In Situ Customization
500-Gene New Panel
350-Gene New Panel
150-Gene Mix-in
50-Gene Add-on
2-Protein Add On
In Situ Workflow
5-Day Sample-to-Answer
Novel Tissue Transfer
Precision Tooling
Parallelization for Simple 4 Flow Cell Prep
In Situ Run Throughput
1 to 4 Flow Cells
2 to 16 Lanes
4 to 128 Samples
4 to 40 cm2
In Situ Analysis
Multimodal Viewer
3rd-Party Compatible Outputs
Clustering and cell segmentation are done onboard, resulting in single-cell sample metrics post-run. Cell segmentation is currently based on nuclear expansion. Membrane-based segmentation is in development and coming soon.
Single Cell Output ?
Note: G4X is currently in development. Specifications, configurations, and capabilities are subject to change. Please contact us for the latest updates on availability and performance.
Spatial Reagents
Flow Cell
Configuration
Throughput
Run Time1
X2s
2 Lanes
10x10 mm ROIs
4 Samples
4 cm2
~24 Hours
X2
2 Lanes
10x10 mm ROIs
10 Samples
10 cm2
~29 Hours
X4
4 Lanes
4.5x4.5 mm ROIs
32 Samples
6.5 cm2
~24 Hours
1Spatial runtime does not include analysis time since spatial analysis occurs after run completion. Analysis time depends on number of flow cells and configurations being run. Typically analysis is a bit over 1 day per flow cell.
Note: G4X is currently in development. Specifications, configurations, and capabilities are subject to change. Please contact us for the latest updates on availability and performance.
Note: G4X is currently in development. Specifications, configurations, and capabilities are subject to change. Please contact us for the latest updates on availability and performance.
Sequencing Reagents
Flow Cell
Config.1
# REads2
Run Time3
Quality4
F3
100 Cycles
300 Cycles (2x150)
100M / Lane
400M / FC
1.6B / Run
12-24 Hours
85% ≥ Q30
1Reagents include 50 additional cycles above what is represented to account for index sequencing needs.
2Indicates typical paired reads passing filter. Throughput may vary depending on application and read length.
3Sequencing run time includes analysis time, and is measured from run start through clustering, sequencing, and instrument wash for non-indexed reads. Run time will vary depending on the number of flow cells and the high-performance computing configuration.
4Performance metrics may be impacted by application, sample quality, library preparation, loading concentration, and other sequencing considerations. Metrics as generated on Salmonella and PhiX control libraries.
Note: G4X is currently in development. Specifications, configurations, and capabilities are subject to change. Please contact us for the latest updates on availability and performance.
2Indicates typical paired reads passing filter. Throughput may vary depending on application and read length.
3Sequencing run time includes analysis time, and is measured from run start through clustering, sequencing, and instrument wash for non-indexed reads. Run time will vary depending on the number of flow cells and the high-performance computing configuration.
4Performance metrics may be impacted by application, sample quality, library preparation, loading concentration, and other sequencing considerations. Metrics as generated on Salmonella and PhiX control libraries.
Note: G4X is currently in development. Specifications, configurations, and capabilities are subject to change. Please contact us for the latest updates on availability and performance.
Learn More
Speak to an Expert
Schedule a seminar or simply get your questions answered.