Spatial Multimodal Analysis on G4X
Powering deep single-cell insights from unified spatial omics and advanced analytical pipelines.
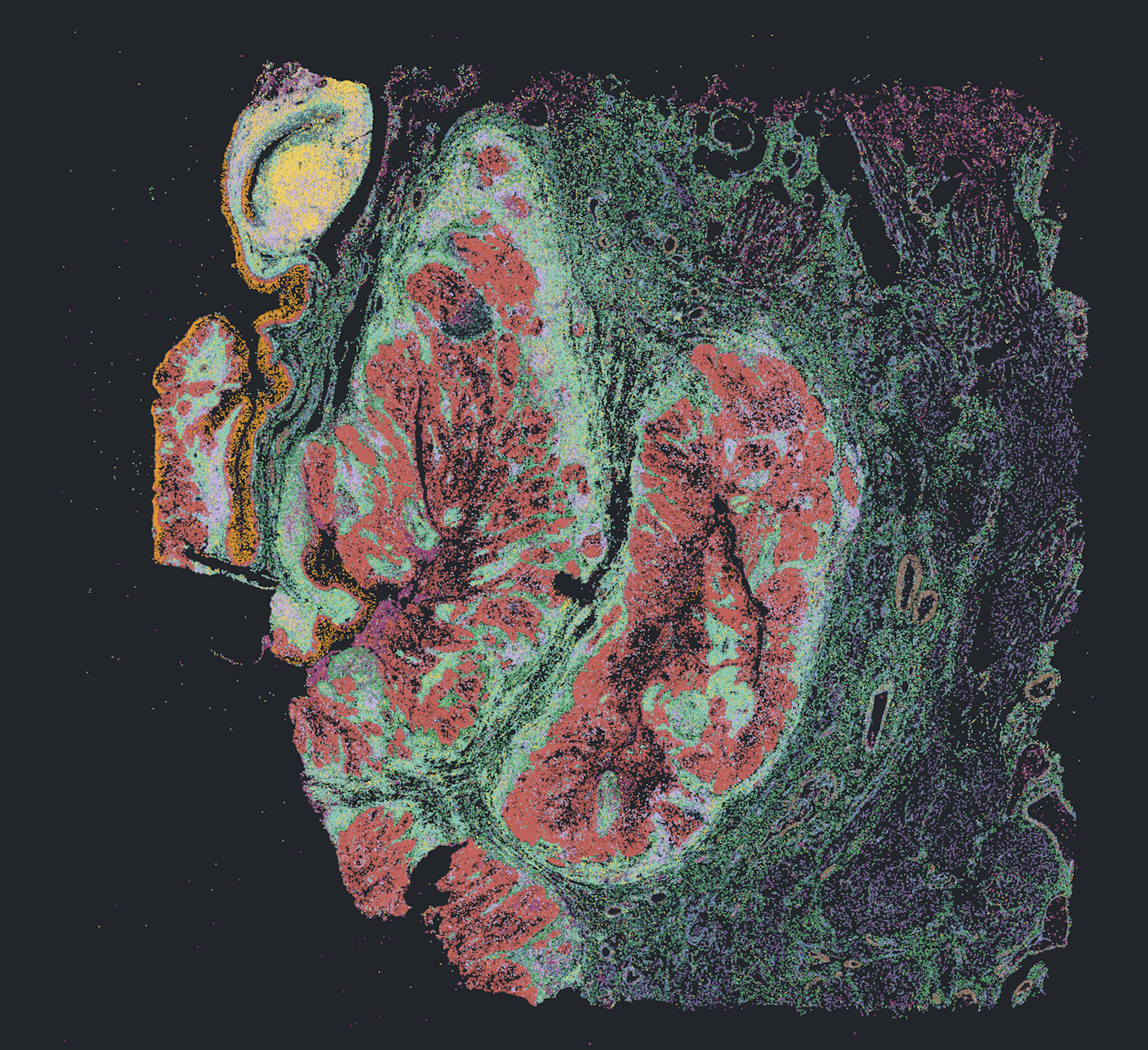
3D niches from multiomic reconstruction of 10 serial renal cell carcinoma sections on single G4X flowcell (6.2 M cells).
Primary Analysis and Results QC
On-board Primary Analysis and QC
High performance computing on the G4X performs image processing, base calling, and machine learning–based single-cell segmentation, eliminating the need for external compute infrastructure and accelerating your time to results.
Viewer
Multimodal Tissue Viewer
High performance computing on the G4X performs image processing, base calling, and machine learning–based single-cell segmentation, eliminating the need for external compute infrastructure and accelerating your time to results.
Viewer
Multimodal Tissue Viewer
High performance computing on the G4X performs image processing, base calling, and machine learning–based single-cell segmentation, eliminating the need for external compute infrastructure and accelerating your time to results.
Demo Data
Analyze yourself
Robust chemistry, precisely controlled workflow, and inherent system throughput enable exceptional section-to-section reproducibility within a lane, across flow cells and across runs. This consistency ensures reliable and comparable results across multiple experiments and samples, facilitating robust spatial mulitiomic analysis

Pearson correlation of per-probe expression levels with positive probes (green) and negative controls between serial sections of kidney cancer FFPE sample profiled on two separate flow cells.
Continue Exploring
View the Poster
AGBT 2025
In-situ multiomic profiling of tumor-immune interactions in 2D and 3D on the G4X Spatial Sequencer
Kenneth G. et al
Download the Datasets
Explore the data yourself using your preferred pipeline.
Check the Panel Content
See the gene targets for our off-the-shelf panels.
Speak to an Expert
Schedule a technical seminar and get your questions answered.



